Cryo-Electron Microscopy of Viral Infection
Prof. Dr. Petr Chlanda
We are interested in studying how viruses interact with cellular organelles and how they remodeled cells to replicate. Many enveloped viruses such as influenza, SARS-CoV-2 or Ebola must cross the cell membrane during entry and exit. To do so, viruses developed or hijacked different protein machinery, which is able to remodel, fuse or cut membranes in processes dependent on specialized lipids. We use and further develop cryo-electron microscopy (cryo-EM) techniques such as cryo-focused ion beam milling (cryo-FIB), in situ cryo-electron tomography (cryo-ET) and cryo-correlative light and electron microscopy (cryo-CLEM) in conjunction with other imaging methods as fluorescence microscopy and imaging mass spectrometry to solve the puzzle of viral-membrane interactions.
“On a molecular level, virus-infected cell is a vast unexplored universe whose understanding will not be possible without constant effort to advance imaging technologies.”
Research Strategy
We aim to understand the molecular mechanisms underlying the membrane and cellular remodelling processes that are pivotal in the viral infection of RNA membrane-enveloped viruses. Influenza A virus, SARS-CoV-2, or the Ebola virus remodel cells to replicate and evade the immune system and they must cross the cell membrane during entry and exit. Viruses have developed or hijacked different protein machinery, which is able to remodel, fuse, or cut membranes in processes dependent on specialised lipids. We use and further develop cryo-electron microscopy (cryo-EM) techniques such as cryo-focused ion beam milling (cryo-FIB), in situ cryo-electron tomography (cryo-ET), and cryo-correlative light and electron microscopy (cryo-CLEM) in conjunction with other imaging methods, such as fluorescence microscopy and imaging mass spectrometry, to solve the puzzle of viral-membrane interactions.
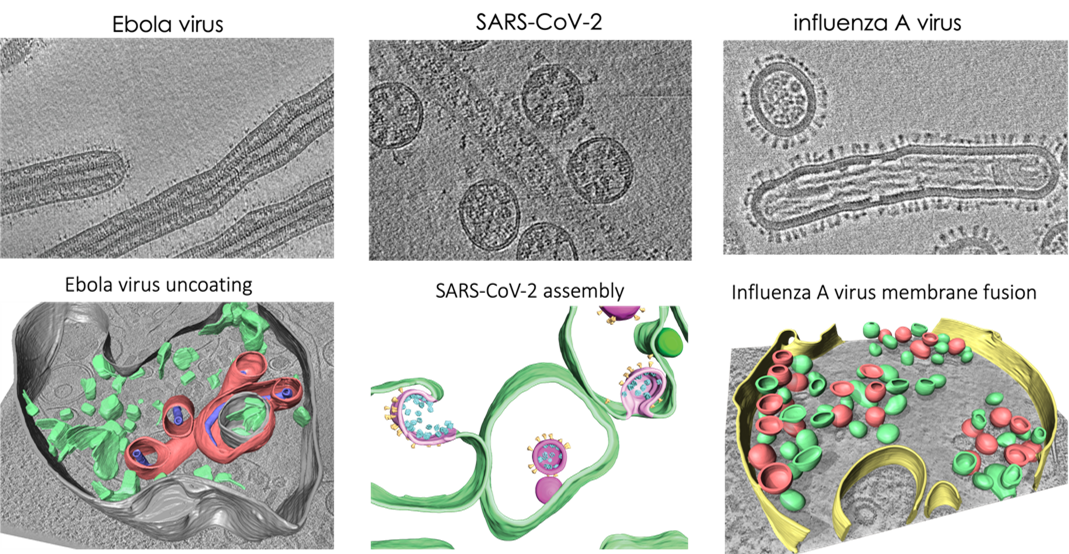
In particular, we study membrane fusion and entry of spherical and filamentous influenza A viruses and we are interested in how influenza A viruses incorporate segmented genomes into the budding viruses. We study replication organelles and assembly of SARS-CoV-2 and Ebola viruses.

Prof. Dr. Petr Chlanda
- +49 (0)6221 54-51231
- petr.chlanda@bioquant.uni-heidelberg.de
Administration
Patcharaporn Rathgeber (Mrs.)
- +49 6221 56-39747
- patcharaporn.rathgeber@med.uni-heidelberg.de
Selected Publications
Nucleocapsid assembly drives Ebola viral factory maturation and dispersion
Melina Vallbracht, Bianca S Bodmer, Konstantin Fischer, Jana Makroczyova, Sophie L Winter, Lisa Wendt, Moritz Wachsmuth-Melm, Thomas Hoenen, Petr Chlanda
Cell 2025 Feb 6;188(3):704-720.e17. doi: 10.1016/j.cell.2024.11.024.
SARS-CoV-2 nsp3 and nsp4 are minimal constituents of a pore spanning replication organelle.
Zimmermann L, Zhao X, Makroczyova J, Wachsmuth-Melm M, Prasad V, Hensel Z, Bartenschlager R, Chlanda P.
Nat Commun. 2023 Nov 30;14(1):7894. doi: 10.1038/s41467-023-43666-5.
The Ebola virus VP40 matrix layer undergoes endosomal disassembly essential for membrane fusion.
Winter SL, Golani G, Lolicato F, Vallbracht M, Thiyagarajah K, Ahmed SS, Lüchtenborg C, Fackler OT, Brügger B, Hoenen T, Nickel W, Schwarz US, Chlanda P.
EMBO J. 2023 Jun 1;42(11):e113578. doi: 10.15252/embj.2023113578.
IFITM3 blocks influenza virus entry by sorting lipids and stabilizing hemifusion.
Klein S, Golani G, Lolicato F, Lahr C, Beyer D, Herrmann A, Wachsmuth-Melm M, Reddmann N, Brecht R, Hosseinzadeh M, Kolovou A, Makroczyova J, Peterl S, Schorb M, Schwab Y, Brügger B, Nickel W, Schwarz US, Chlanda P.
Cell Host Microbe. 2023 Apr 12;31(4):616-633.e20. doi: 10.1016/j.chom.2023.03.005.
SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography.
Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P.
Nat Commun. 2020 Nov 18;11(1):5885. doi: 10.1038/s41467-020-19619-7.
Full publication list of the group leader on PubMed